By Frank Kuo
This post originally appeared on CLEO 2011 by Frank Kuo and is reproduced with permission from its author.
Recently, three-dimensional plasmon rulers based on nano-rods are reported on Science. Hopefully, it will be cool weaponry for measuring the structures of the molecules in the near future. Over the past few decades, optical rulers based on different principles emerged from various branches of optical science. At the same time, researchers are trying hard to push each methodology to the limit. Optical Rulers, as a result, attract an army of researchers and spin off fruitful results. A quick summary of them seems to be a fair amount of content for everyone.
First thing first, what does an optical ruler do? Quite straightforward, it measures the dimensions of the molecular structures. For example, what is the distance between two subunits of a hemoglobin protein? What is the height of a membrane protein when measured from the membrane surface? Put into one sentence – optical rulers are aimed to map out the 3D structures of the molecules such that we can use this information to figure out the functionality of the molecules.
First ruler comes to your mind, I guess, will be the technique of X-ray diffraction. It is a true powerful optical ruler. After all, the DNA structure is solved by it, and Nobel Prize acclaims this technique for more than once. It has great resolution ~ 1Å, and you do not have put anything attached to the molecules. However, the pitfall is that, you have to crystallize the molecules which are merely impossible for some molecules, and the crystal forms of the molecules are in general, not the in vivo forms of the molecules. A report on C&EN and JACS beautifully illustrate the structure changes dramatically depending on the environment.
What is the king of in vivo optical ruler? I would say so far it is NMR. It has the resolutions of a few Å, and the algorithm is advanced so much that complex proteins are revealing their true forms (through more advanced multi-dimensional NMR). The principle is very similar to the trick we play with tuning forks. If you hit on one of the forks and bring the other replica close in, you feel the vibration on the second one and actually both will make the same tone without two touching each other. The energy (in terms of sound wave) resonates in these two forks. In NMR, intrinsic atomic spin plays the role of the tuning fork. By incorporate the isotopes (such as 1H 13C and 15N, these atoms have nonzero spins) into the amino acids of the proteins, the spins of these atoms behave like tuning forks with different tones. Imaging if there are two or more isotope atoms close to each other, the energy (in this situation, the microwave qunta) will be transferred between the isotopes (let’s say between 1H and 15N) assuming one of them is excited by microwave. NMR is specialized in measuring this energy transfer. The closer they are the more efficient energy transfer is. This efficiency drops proportional to 1/r^-6, where r is the distance between two spins. Now, if you have many of these isotope atoms located at different amino acids of a protein, you can figure out which isotopes (or more interestingly, which amino acids) are closer to each other. With the help of computer, you can infer the structures of the proteins, much like a complex trigonometry based on the relative positions of the spins.
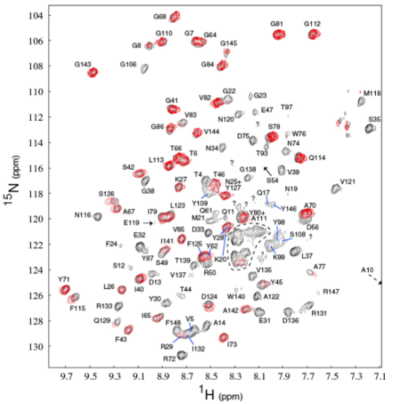 Figure 1. A typical 2D NMR spectrum. Each blob can be thought as a sign of energy transferring between the spins of some specific hydrogen and nitrogen. By doing this kind of cross mapping, we can figure out the 3D structure of the molecules.
Figure 1. A typical 2D NMR spectrum. Each blob can be thought as a sign of energy transferring between the spins of some specific hydrogen and nitrogen. By doing this kind of cross mapping, we can figure out the 3D structure of the molecules.
Continue reading »
Posted: 28 June 2011 by
Frank Kuo
| with 0 comments